Google AI’s Five-Year Triumph: AlphaFold Sweeps Nobel Prize, Predicts 200M Protein Structures
AlphaFold: AI Smashes a 50-Year Protein Structure Puzzle
Xinzhiyuan Report
[Xinzhiyuan Lead] — AI has compressed a decades-long challenge into minutes: AlphaFold is now used by 3.3 million researchers worldwide. In Turkey, two undergraduates have harnessed this free tool to complete 15 structural studies, dismantling barriers to entry in science. For the first time, research is pacing at digital speed.
---
Late Nights in the Lab vs. AI in the Browser
Have you ever worked in the lab until 2 a.m.?
Tasks pile up: enzyme cutting, purification, sample loading, parameter tuning… finally, staring at faint bands and wondering:
If this doesn’t work, my year's effort is gone.
Meanwhile, on the other side of the globe: two Turkish undergraduates are staring at a screen — waiting not for instrument readouts, but for an AI-generated protein structure diagram from AlphaFold.
Within months, they compiled 15 papers from these structures — all via a free webpage, without top-tier mentors or million-dollar equipment.
This is an efficiency revolution and a super tool manual for the next generation.

---
A 50-Year Puzzle Solved in Minutes by AI
The question: What exactly does a protein look like?
- Proteins are microscopic machines inside cells.
- Made of amino acid chains, they must fold precisely into a 3D structure.
- Even slight misfolding can cause malfunction or disease.
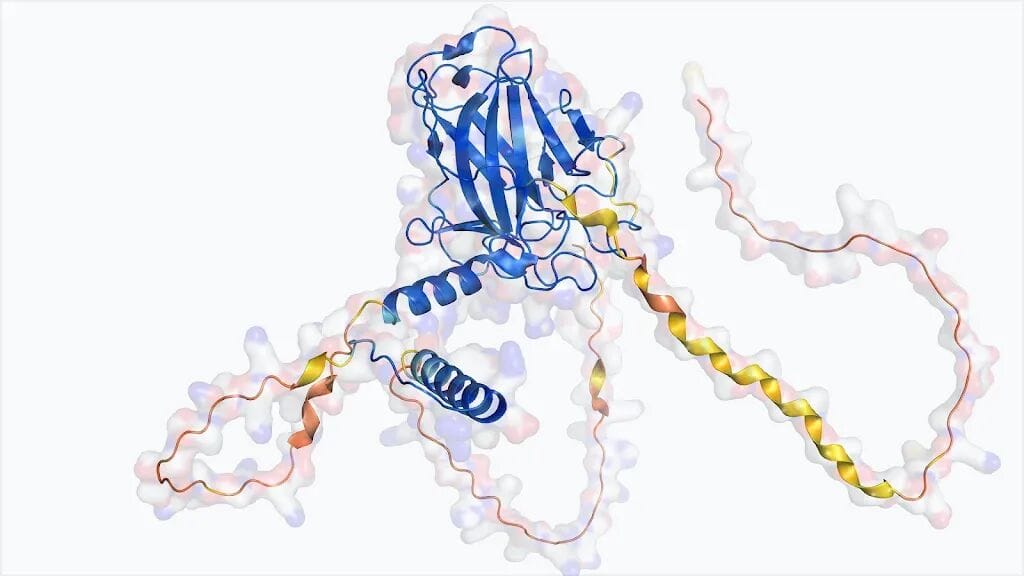
AlphaFold model of p53, a key cancer-associated protein.
Traditional method:
- Time: 1+ years per protein
- Cost: millions in experiments
- Techniques: X-ray crystallography, cryo-EM
- Result: long queues and stalled research timelines
Breakthrough: In 2020 at CASP14, AlphaFold 2 predicted protein structures from sequences with accuracy comparable to experiments.
From then, experiments were no longer the only gateway.

---
A Free Webpage Unlocks the “Black Box”
DeepMind released AlphaFold 2’s code + predicted structures free online.
The AlphaFold Protein Structure Database now holds 200M+ structures.

- Producing this volume experimentally would take millions of years.
- Nature: database accessed by 3.3M users in 190+ countries; 1M from low/mid-income regions.
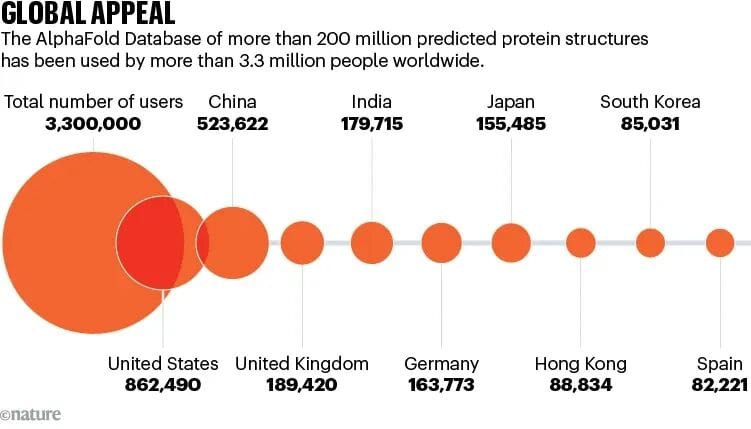
Impact: Structural biology now has global reach. Anyone can explore proteins at atomic detail — even undergraduates.
Case: EAAT1 Membrane Protein
- Located on brain cell membranes, transports neurotransmitters.
- Traditionally, membrane proteins are extremely hard to characterize experimentally.
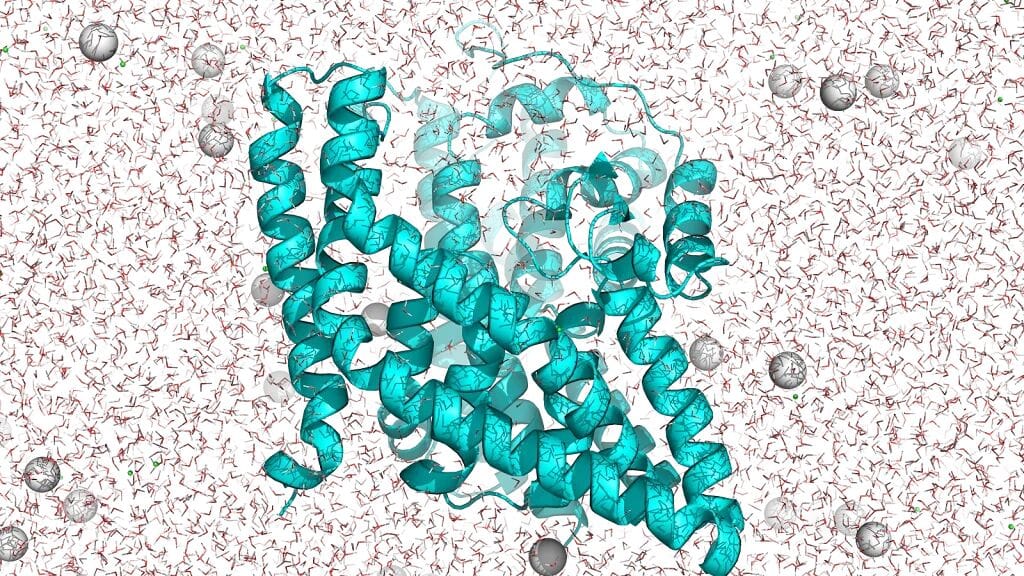
With AlphaFold, they virtually disassembled and reconstructed EAAT1 in 3D.
They also used the QTY method — replacing hydrophobic amino acids to make the protein easier to work with experimentally.
---
Global Trend: From Validation to Exploration
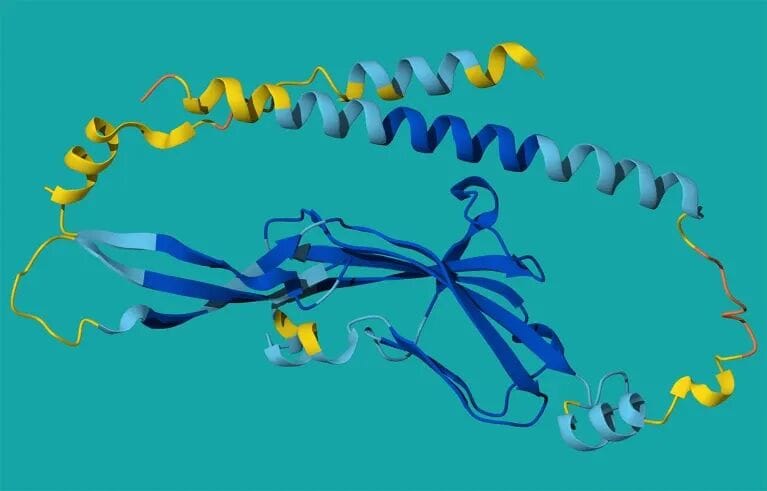
Bouncer–Tmem81 Complex predicted by AlphaFold, explaining how sperm recognizes eggs in zebrafish.
Examples from Nature:
- apoB100 — key in atherosclerosis, untangled for the first time
- p53 — vital cancer target
- AlphaFold users submit 40% more novel structures than non-users
- In Protein Data Bank, AlphaFold teams submit 50% more structures
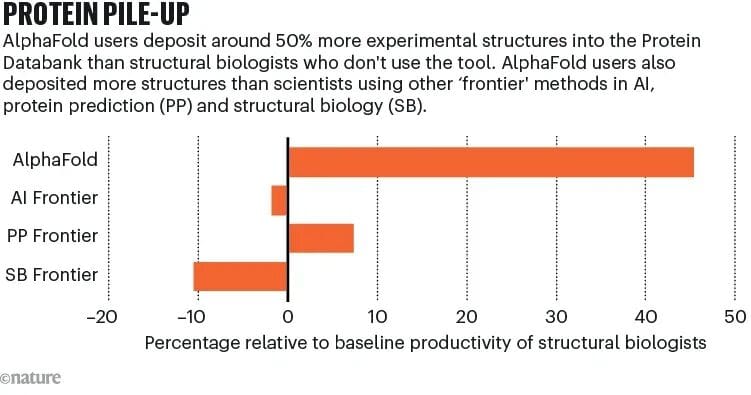
Quote from Nature:
> Structural biology is shifting from “validation territory” to “exploration territory.”
---
Citation & Impact
- AlphaFold2’s Nature paper citation rate still rising — ~800/year average.
- Linked studies twice as likely to be cited in clinical papers.
- Significantly higher likelihood of patent citations.
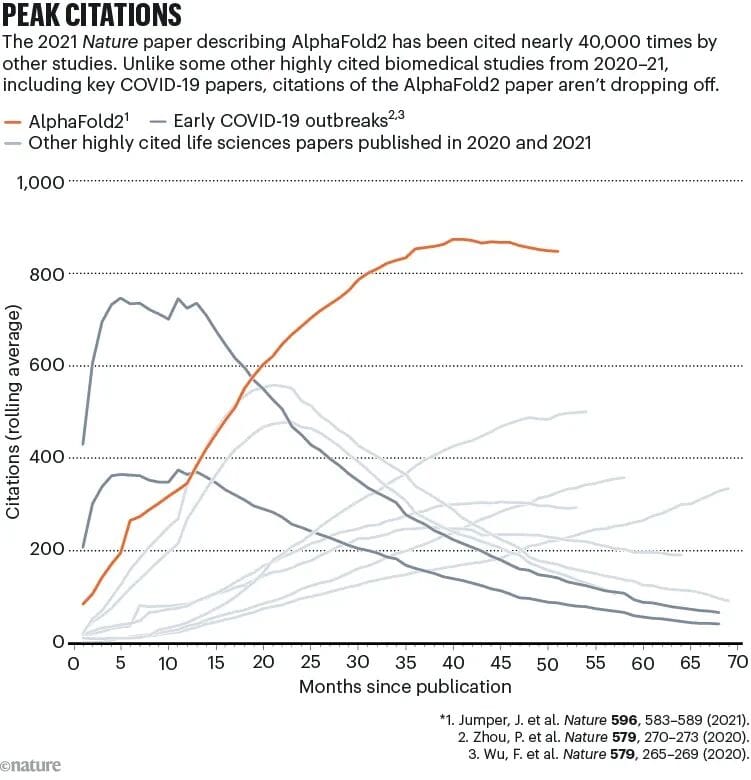
This is not a “one-time hype”; it’s a continuously reused tool in translational science.
---
AlphaFold 3: From Structure to Interaction
AlphaFold2 answered “What does a folded protein look like?”
AlphaFold3 asks “How do life’s molecules interact?”
Capabilities
- Predict protein–DNA/RNA, protein–ligand, and glycan interactions in 3D
- Visualize candidate drugs fitting into binding pockets
- Model virus spike proteins with sugar shields recognized by antibodies

---
Applications: From Labs to Farms
- Isomorphic Labs: AlphaFold3 in AI-powered drug discovery
- Bees: decoding Vitellogenin for disease resistance breeding
- Plants: accelerating environmental response protein research


---
The New Scientific Workflow
Two researchers:
- In a lab, manually pushing experiments.
- In a browser, AI-handling structural prediction in minutes.
The future favors those who mobilize tools and ask bold questions.
Even without millions in lab gear, you can:
- Open the AlphaFold database
- Type a protein name
- Get an atom-level blueprint — free
The wall between resource-rich labs and ordinary researchers is loosening.
Your position depends on whether you press “Run”.
---
References
- https://deepmind.google/blog/alphafold-five-years-of-impact/
- https://www.nature.com/articles/d41586-025-03886-9
---
Amplifying Science with AiToEarn
Platforms like AiToEarn官网 extend AlphaFold’s ethos into science communication:
- AI content generation
- Cross-platform publishing (Douyin, Kwai, WeChat, Bilibili, Xiaohongshu, Facebook, Instagram, LinkedIn, Threads, YouTube, Pinterest, X/Twitter)
- Analytics & AI model ranking (AI模型排名)
Together, such tools help ensure breakthroughs reach the widest possible audience efficiently — as seamlessly as AlphaFold leaps from sequence to structure.
---
Would you like me to also create a short infographic-style summary version of this that highlights the most striking stats and visuals for quick sharing? That could greatly enhance engagement for broader audiences.



